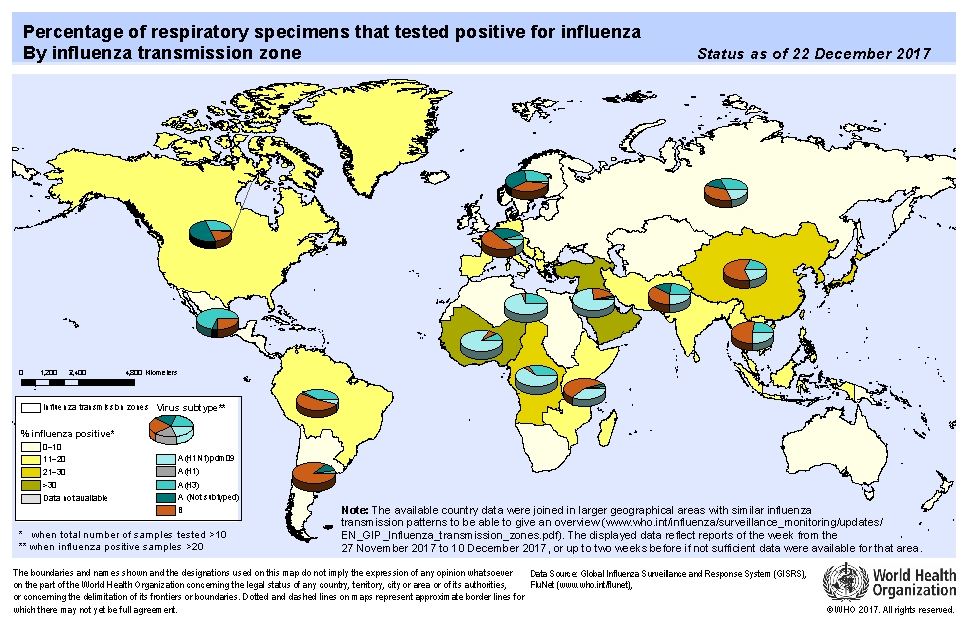
1. GLOBAL WHO SUMMARY – based on data up to Dec 22th 2017
North America: overall activity continued to increase in the region, with detections of predominantly influenza A(H3N2) viruses.
Europe: activity continued to increase, but remained low in most of the countries, with detections of predominantly influenza B followed by influenza A(H3N2) viruses.
Asia: Western Asia: elevated levels of activity were reported in in recent weeks, with influenza A(H1N1)pdm09 predominantly detected. Central Asia: low to no activity was reported. East Asia: influenza activity remained low in most of the countries with the exception of China where influenza like illness (ILI) and influenza percentage positive continued to increase, with influenza B/Yamagata viruses predominantly detected. South East Asia: low levels of influenza activity were reported. Southern Asia: influenza activity remained low in general. Detections of influenza A(H1N1)pdm09 and A(H3N2) viruses were reported in India and of all seasonal subtypes in the Islamic Republic of Iran.
Africa: Northern Africa: low levels of influenza activity were reported. Detections of influenza A(H1N1)pdm09 virus increased slightly in Tunisia. Middle Africa: sporadic detections of influenza A were reported in Cameroon. Eastern Africa: influenza A(H3N2) and B detections were reported in Madagascar and Mozambique.
In the Caribbean and Central American countries: respiratory illness indicators and influenza activity remained low in general but respiratory syncytial virus (RSV) activity remained high in several countries.
2. VIROLOGICAL SURVEILLANCE AND STRAIN CHARACTERIZATION
WHO GISRS laboratories: from 27 November 2017 to 10 December 2017
127006 specimens were tested during that time period. 15344 (12,0%) were positive for influenza viruses, of which 9579 (62.4%) were typed as influenza A and 5765 (37.6%) as influenza B. Of the sub-typed influenza A viruses, 1596 (30.1%) were influenza A(H1N1)pdm09 and 3698 (69.9%) were influenza A(H3N2). Of the characterized B viruses, 2640 (85.2%) belonged to the B-Yamagata lineage and 460 (14.8%) to the B-Victoria lineage.
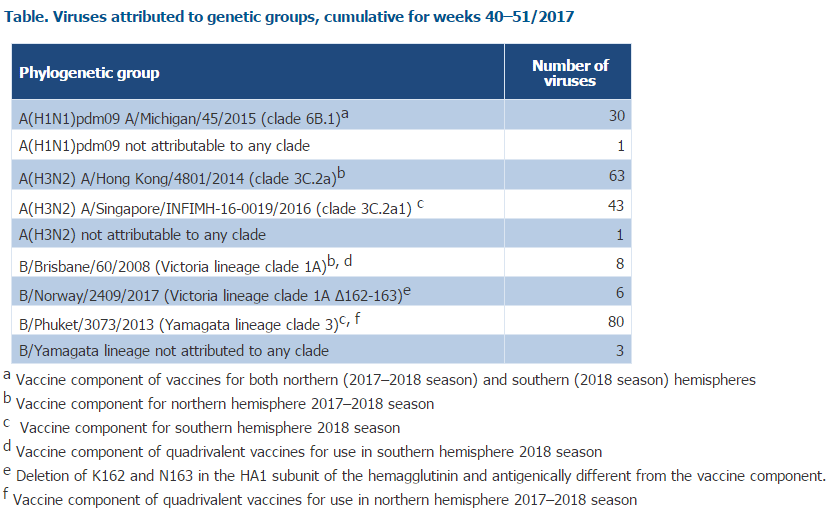
NB: The recommended components for the 2017-2018 Northern hemisphere Influenza vaccine includes: an A/Michigan/45/2015 (H1N1)pdm09-like virus; an A/Hong Kong/4801/2014 (H3N2)-like virus; a B/Brisbane/60/2008-like virus (Vic Lineage). and a B/Phuket/3073/2013-like virus (Yam Lineage) in QIV vaccine.
EUROPE weeks: Week 40 to 51/2017
First detections indicated circulation of A(H3N2) and B/Yamagata viruses in the highest proportions. As the A(H3N2) subtype dominated last season, a high proportion of the population should be protected. For type B viruses from both sentinel and non-sentinel sources, B/Yamagata lineage viruses have greatly outnumbered those of the B/Victoria lineage.
While low in number, 59% of the genetically characterized A(H3N2) viruses belonged to clade 3C.2a, the vaccine virus clade as described in the WHO recommendations for vaccine composition for the NH 2017–18, and 40% to clade 3C.2a1, the viruses of which are antigenically similar to those of clade 3C.2a.
For specimens collected since week 40/2017, genetic characterization of 235 viruses has been reported. Among 107 influenza A(H3N2) viruses, 63 (59%) fell in the vaccine virus component clade (3C.2a), and 42 (40%) in subclade 3C.2a1 with viruses defined by N171K, often with N121K, amino acid substitutions in the haemagglutinin. Viruses in these 2 groups are antigenically similar, but both clade and subclade are evolving rapidly with the emergence of several virus clusters defined by additional amino acid substitutions in the haemagglutinin, thereby requiring continued monitoring of antigenic characteristics. 1 A(H1N1)pdm09, 1 A(H3N2) and 3 B/Yamagata viruses were not attributed to any clade.
See also the November report from ECDC.
US-CDC- Until Dec 23th, 2017
The most frequently identified influenza virus subtype reported by public health laboratories during week 51 was influenza A(H3). The percentage of respiratory specimens testing positive for influenza in clinical laboratories increased.
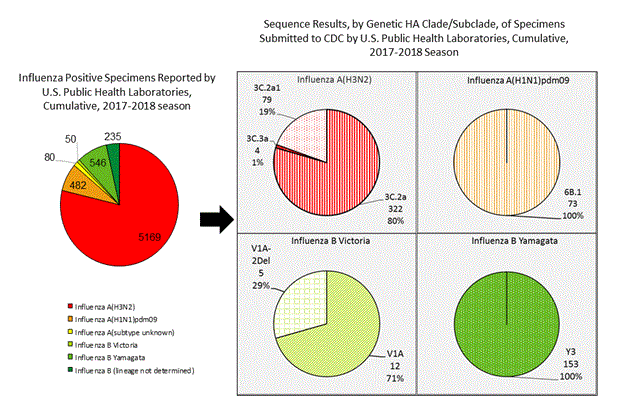
CDC has antigenically or genetically characterized 648 influenza viruses collected during October 1 – December 23, 2017 including 73 influenza A(H1N1)pdm09 viruses, 405 influenza A(H3N2) viruses, and 170 influenza B viruses.
A (H1N1)pdm09: Phylogenetic analysis of the HA genes from 73 A(H1N1)pdm09 viruses showed that all belonged to clade 6B.1. Forty-one A(H1N1)pdm09 viruses were antigenically characterized, and all were antigenically similar (analyzed using HI with ferret antisera) to the reference 6B.1 virus A/Michigan/45/2015, representing the recommended influenza A(H1N1)pdm09 reference virus for the 2017–18 NH vaccines.
A (H3N2): Phylogenetic analysis of the HA genes from 405 A(H3N2) viruses revealed extensive genetic diversity with multiple clades/subclades co-circulating. The HA genes of circulating viruses belonged to clade 3C.2a (n=322), subclade 3C.2a1 (n=79) or clade 3C.3a (n=4). One hundred twenty seven influenza A(H3N2) viruses were antigenically characterized, and 126 (99.2%) A(H3N2) viruses tested were well-inhibited (reacting at titers that were within fourfold of the homologous virus titer) by ferret antisera raised against A/Michigan/15/2014 (3C.2a), a cell propagated A/Hong Kong/4801/2014-like reference virus representing the A(H3N2) component of 2017–18 NH vaccines.
B/Victoria: Phylogenetic analysis of 17 B/Victoria-lineage viruses indicate that all HA genes belonged to genetic clade V1A, the same genetic clade as the vaccine reference virus, B/Brisbane/60/2008. However, a small number of viruses identified in 2017 had a 6-nucleotide deletion (encoding amino acids 162 and 163) in the HA (abbreviated as V1A-2Del). Four (57.1%) B/Victoria lineage viruses were well-inhibited by ferret antisera raised against cell -propagated B/Brisbane/60/2008 reference virus, representing a recommended B virus component of 2017–18 NH vaccines. Three (42.9%) B/Victoria lineage virus reacted poorly (at titers that were 8-fold or greater reduced compared with the homologous virus titer) with ferret antisera raised against cell-propagated B/Brisbane/60/2008, and these viruses had the V1A-2Del HA.
B/Yamagata: Phylogenetic analysis of 153 influenza B/Yamagata-lineage viruses indicate that the HA genes belonged to clade Y3. A total of 71 influenza B/Yamagata-lineage viruses were antigenically characterized, and all were antigenically similar to cell propagated B/Phuket/3073/2013, the reference vaccine virus representing the influenza B/Yamagata-lineage component of the 2017–18 NH QIV.
HPA-London until Week 01,2018
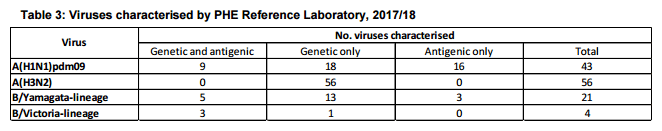
The PHE Respiratory Virus Unit has characterised 124 influenza viruses detected since week 37. Of the 43 A(H1N1)pdm09 influenza viruses that have been characterised, all belong in the genetic subgroup 6B.1, which was the predominant genetic subgroup in the 2016/17 season and to date during the current season. The 25 viruses antigenically analysed are similar to the A/Michigan/45/2015 NH 2017/18 (H1N1)pdm09 vaccine strain.
Genetic characterisation of 56 A(H3N2) influenza viruses detected since late summer, showed that they all belong to genetic subclade 3C.2a, with 32 belonging to a cluster within this genetic subclade designated as 3C.2a1. The NH 2017/18 influenza A(H3N2) vaccine strain A/HongKong/4801/2014 belongs in genetic subclade 3C.2a.
Twenty five influenza B viruses have been analysed; 21 were characterised as belonging to the B/Yamagata/16/88-lineage and 4 belonging to the B/Victoria/2/1987-lineage. Of the influenza B viruses antigenically characterised, the B/Victoria/2/87-lineage viruses were antigenically similar to B/Brisbane/60/2008, the influenza B/Victoria-lineage component of 2017/18 NH TIV trivalent and QIV. B/Yamagata/16/88-lineage viruses were antigenically similar to B/Phuket/3073/2013, the influenza B/Yamagata-lineage component of the 2017/18 NH QIV vaccine.
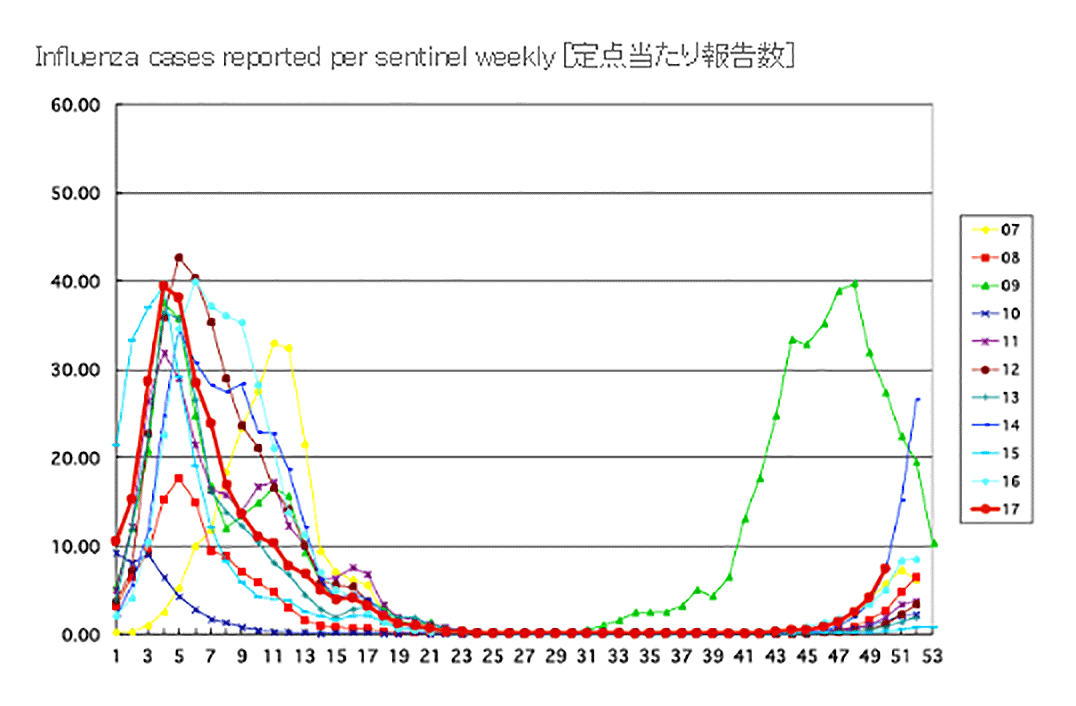
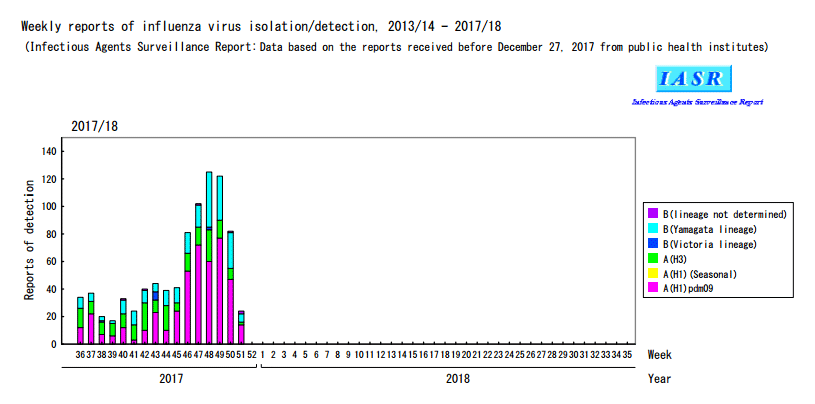
JAPAN National infectious diseases- Until Week 51/2017
3. MORE DATA FROM REGIONAL OR COUNTRIES SURVEILLANCE
- Canada
Government, FluWatch Report - China
Chinese National Influenza Center – Influenza Weekly Report- Week - Europe
ECDC/ WHO Europe Weekly Influenza update - Hong Kong
Centre for Health Protection- Flux Express Surveillance Report - PAHO
Regional Update - Russian Federation
WHO NIC at Research Institute of Influenza and D.I Ivanovsky Institute of Virology, Integrated data of influenza morbidity and diagnosis - Singapore
Ministry of Health – weekly infectious disease bulletin - Unites States Of America
Centers for Disease Control and Prevention, Weekly U.S. Influenza Surveillance Report

buying prescription drugs in mexico online: cheapest mexico drugs – mexico pharmacies prescription drugs
india pharmacy mail order http://indiaph24.store/# cheapest online pharmacy india
top 10 online pharmacy in india
mexican pharmacy: cheapest mexico drugs – medicine in mexico pharmacies
online canadian pharmacy reviews Large Selection of Medications from Canada best online canadian pharmacy
https://nolvadex.life/# tamoxifen men
cost of generic propecia without a prescription: cost generic propecia price – get cheap propecia without dr prescription
cheap propecia pill cost of generic propecia price propecia sale
http://nolvadex.life/# tamoxifen hot flashes
buy cytotec pills online cheap: Cytotec 200mcg price – Cytotec 200mcg price
http://finasteride.store/# order propecia without rx
https://cytotec.club/# cytotec online
cost propecia tablets cost cheap propecia for sale propecia without insurance
effexor and tamoxifen: liquid tamoxifen – tamoxifen endometriosis
propecia order generic propecia pill order cheap propecia without dr prescription
http://ciprofloxacin.tech/# cipro
lisinopril 5 mg canada: lisinopril 5mg buy – lisinopril 20 mg prices
http://lisinopril.network/# cost of lisinopril 40 mg
how to prevent hair loss while on tamoxifen tamoxifen menopause tamoxifen skin changes
http://nolvadex.life/# common side effects of tamoxifen
order propecia without insurance: generic propecia without insurance – order generic propecia for sale
buy cheap lisinopril 40mg zestoretic coupon lisinopril cost
https://ciprofloxacin.tech/# buy cipro online without prescription
cipro online no prescription in the usa: ciprofloxacin 500mg buy online – buy ciprofloxacin over the counter
buy cytotec over the counter п»їcytotec pills online buy cytotec online fast delivery
https://ciprofloxacin.tech/# ciprofloxacin 500 mg tablet price
http://ciprofloxacin.tech/# ciprofloxacin mail online
tamoxifen bone pain femara vs tamoxifen tamoxifen for breast cancer prevention
buy cytotec in usa: cytotec buy online usa – cytotec buy online usa
http://lisinopril.network/# lisinopril 10mg tablets
Abortion pills online buy cytotec online fast delivery buy cytotec pills online cheap
cipro for sale: cipro 500mg best prices – buy ciprofloxacin over the counter
https://cytotec.club/# buy cytotec pills online cheap
http://lisinopril.network/# lisinopril 20mg pill
buy cytotec over the counter cytotec pills buy online buy cytotec in usa
effexor and tamoxifen: tamoxifen rash – nolvadex generic
buy cheap propecia buy generic propecia no prescription cost generic propecia pill
http://lisinopril.network/# lisinopril 3
A lot of thanks for each of your effort on this web site. My mom enjoys carrying out research and it is easy to understand why. Many of us notice all of the powerful mode you give effective information on your website and in addition recommend response from some others on that content and our own child is truly understanding a lot. Take pleasure in the rest of the year. You have been performing a tremendous job.
tamoxifen rash: pct nolvadex – where to get nolvadex
buying propecia for sale buy cheap propecia online cost of propecia
https://cytotec.club/# buy cytotec in usa
http://finasteride.store/# get cheap propecia without insurance
zestril 10 mg in india: price of lisinopril generic – rx lisinopril
price of zestril 30 mg lisinopril 60 mg tablet no prescription lisinopril
generic sildenafil: viagras.online – viagra without prescription
http://cialist.pro/# Buy Tadalafil 10mg
buy cenforce: order cenforce – cenforce.pro
Buy Cenforce 100mg Online Cenforce 150 mg online buy cenforce
http://cialist.pro/# Generic Tadalafil 20mg price
https://cialist.pro/# Cialis without a doctor prescription
Cheap Levitra online: buy Levitra over the counter – Buy Vardenafil 20mg online
Tadalafil price Tadalafil Tablet Cialis 20mg price in USA
Levitra online pharmacy: Buy Vardenafil 20mg – Buy Vardenafil 20mg online
http://viagras.online/# Sildenafil 100mg price
buy Levitra over the counter: Cheap Levitra online – Vardenafil buy online
sildenafil oral jelly 100mg kamagra kamagra.win sildenafil oral jelly 100mg kamagra
http://kamagra.win/# п»їkamagra
buy cialis pill Cialis 20mg price in USA Cheap Cialis
https://levitrav.store/# Buy Vardenafil online
order cenforce: order cenforce – buy cenforce
http://viagras.online/# Viagra online price
Buy Viagra online cheap Cheapest Sildenafil online viagra canada
http://viagras.online/# Cheap generic Viagra
Tadalafil Tablet: cialist.pro – Generic Cialis price
Viagra without a doctor prescription Canada Buy generic 100mg Viagra online Cheap generic Viagra online
http://levitrav.store/# Levitra tablet price
Buy Cialis online: buy cialis overseas – Cialis 20mg price
canadian pharmacy online no prescription needed: canadian rx prescription drugstore – buy medication online with prescription
http://pharmcanada.shop/# canadian online pharmacy reviews
mexican pharmaceuticals online: buying prescription drugs in mexico online – mexican border pharmacies shipping to usa
buying online prescription drugs canadian pharmacy without prescription pharmacy online no prescription
mexico drug stores pharmacies: mexico pharmacies prescription drugs – mexico drug stores pharmacies
http://pharmnoprescription.icu/# buy medication online no prescription
https://pharmcanada.shop/# canadian drugs
reliable canadian pharmacy reviews: the canadian pharmacy – legit canadian pharmacy
no prescription online pharmacies how to order prescription drugs from canada buy medication online no prescription
india pharmacy: Online medicine order – top online pharmacy india
https://pharmworld.store/# cheapest pharmacy for prescriptions
best canadian pharmacy no prescription: canadian pharmacy without prescription – canadian pharmacy coupon
world pharmacy india reputable indian online pharmacy reputable indian online pharmacy
http://pharmcanada.shop/# canadian pharmacy ltd
ed meds online canada canada drugs online safe reliable canadian pharmacy
mexican online pharmacies prescription drugs: mexico drug stores pharmacies – mexican pharmacy
online pharmacy without prescription: pharm world store – canadian pharmacy world coupon
legal canadian pharmacy online: canadian mail order pharmacy – canadian pharmacy price checker
https://pharmmexico.online/# mexican mail order pharmacies
buying prescription drugs in mexico pharmacies in mexico that ship to usa buying from online mexican pharmacy
no prescription needed pharmacy: online drugs no prescription – can you buy prescription drugs in canada
https://pharmworld.store/# canadian online pharmacy no prescription
mexican border pharmacies shipping to usa: buying from online mexican pharmacy – medicine in mexico pharmacies
п»їbest mexican online pharmacies buying prescription drugs in mexico online best online pharmacies in mexico
doxycycline 100mg online: doxycycline 150 mg – how to order doxycycline
http://gabapentinneurontin.pro/# buy cheap neurontin
buy zithromax: generic zithromax medicine – azithromycin zithromax
doxycycline 150 mg 100mg doxycycline doxycycline 200 mg
zithromax tablets: generic zithromax medicine – how to get zithromax over the counter
prednisone over the counter: generic over the counter prednisone – buy 10 mg prednisone
https://amoxila.pro/# where to buy amoxicillin 500mg
zithromax capsules price: where can i buy zithromax capsules – where can i get zithromax over the counter
generic zithromax 500mg: generic zithromax 500mg – zithromax 250
doxycycline hydrochloride 100mg: doxycycline 100mg – buy doxycycline without prescription uk
10 mg prednisone tablets: prednisone 20mg online – online prednisone
zithromax for sale cheap: zithromax buy online no prescription – zithromax
amoxicillin for sale online can you purchase amoxicillin online order amoxicillin 500mg
https://doxycyclinea.online/# doxycycline hyc 100mg
buy neurontin uk: how to get neurontin – neurontin 400 mg tablets
zithromax over the counter uk generic zithromax 500mg zithromax antibiotic without prescription
http://gabapentinneurontin.pro/# neurontin uk
10mg prednisone daily: prednisone for sale without a prescription – buy prednisone canada
amoxicillin without a doctors prescription: amoxicillin medicine – where can i buy amoxocillin
neurontin price comparison cost of neurontin 100mg gabapentin buy
https://amoxila.pro/# can i buy amoxicillin over the counter in australia
40 mg daily prednisone: canada buy prednisone online – prednisone 4 mg daily
prescription price for neurontin: drug neurontin 20 mg – neurontin 100 mg caps
doxycycline mono doxycycline 100 mg vibramycin 100 mg
https://zithromaxa.store/# generic zithromax over the counter
doxycycline mono: doxycycline monohydrate – generic doxycycline
http://amoxila.pro/# amoxicillin 500 mg cost
buying amoxicillin in mexico: amoxil pharmacy – buy amoxicillin online without prescription
prednisone otc uk: compare prednisone prices – prednisone 5 mg
cost of neurontin neurontin tablets neurontin cost uk
http://doxycyclinea.online/# doxycycline
buy prednisone online uk: generic prednisone tablets – how can i order prednisone
doxycycline 100mg: doxycycline – doxycycline
neurontin 1800 mg neurontin 100mg capsule price neurontin 300 mg price in india
https://amoxila.pro/# price of amoxicillin without insurance
zithromax over the counter uk: zithromax over the counter canada – zithromax 500 mg for sale
doxycycline without prescription doxycycline order online doxycycline mono
where can you buy amoxicillin over the counter: amoxicillin generic – buy amoxicillin 500mg canada
http://zithromaxa.store/# generic zithromax india
neurontin pills for sale: 800mg neurontin – neurontin medicine
buy doxycycline hyclate 100mg without a rx buy doxycycline online without prescription doxycycline 50 mg
buy amoxicillin 500mg uk: amoxicillin 500mg buy online canada – amoxicillin 500 mg capsule
https://prednisoned.online/# 50 mg prednisone tablet
5mg prednisone: prednisone 5mg coupon – where to buy prednisone 20mg no prescription
where to buy prednisone 20mg no prescription prednisone buy can i buy prednisone online in uk
https://zithromaxa.store/# zithromax online usa no prescription
cost of amoxicillin 875 mg: buy cheap amoxicillin – where to get amoxicillin over the counter
neurontin pills: neurontin cap – neurontin 800 mg pill
purchase prednisone 10mg prednisone 60 mg by prednisone w not prescription
https://doxycyclinea.online/# doxycycline hyc 100mg
where to buy zithromax in canada: generic zithromax 500mg – zithromax buy
order doxycycline 100mg without prescription: doxycycline hyclate – buy doxycycline cheap
prednisone tablets india where can i get prednisone over the counter order prednisone with mastercard debit
https://prednisoned.online/# prednisone 20 mg without prescription
zithromax for sale online: zithromax 250 mg australia – buy zithromax online
amoxicillin 500mg prescription: generic for amoxicillin – amoxicillin 200 mg tablet
can you buy amoxicillin over the counter in canada amoxicillin 500 mg cost buying amoxicillin in mexico
http://amoxila.pro/# amoxicillin 500mg price in canada
zithromax 250 mg pill: can you buy zithromax over the counter in mexico – zithromax without prescription
doxycycline online: doxycycline 100mg dogs – doxycycline 100mg capsules
neurontin capsules 300mg brand neurontin 100 mg canada how much is generic neurontin
buying prescription drugs in mexico online: buying from online mexican pharmacy – п»їbest mexican online pharmacies
https://mexicanpharmacy1st.shop/# mexico pharmacy
reputable mexican pharmacies online: mexican online pharmacies prescription drugs – pharmacies in mexico that ship to usa
buying prescription drugs in mexico mexico pharmacy mexican border pharmacies shipping to usa
https://mexicanpharmacy1st.com/# buying prescription drugs in mexico online
http://mexicanpharmacy1st.com/# mexican mail order pharmacies
reputable mexican pharmacies online: mexican drugstore online – mexican online pharmacies prescription drugs
mexico pharmacies prescription drugs mexican online pharmacies prescription drugs mexican border pharmacies shipping to usa
buying prescription drugs in mexico: best online pharmacies in mexico – mexican online pharmacies prescription drugs
https://mexicanpharmacy1st.online/# buying prescription drugs in mexico
buying prescription drugs in mexico: mexican drugstore online – mexico drug stores pharmacies
buying prescription drugs in mexico: buying from online mexican pharmacy – mexican drugstore online
mexican drugstore online buying prescription drugs in mexico online mexico drug stores pharmacies
mexico pharmacies prescription drugs: mexican pharmacy – purple pharmacy mexico price list
https://mexicanpharmacy1st.online/# mexican pharmacy
mexican online pharmacies prescription drugs п»їbest mexican online pharmacies mexico drug stores pharmacies
https://mexicanpharmacy1st.com/# buying prescription drugs in mexico
mexico pharmacies prescription drugs: mexico pharmacies prescription drugs – mexico drug stores pharmacies
mexican rx online: pharmacies in mexico that ship to usa – mexican border pharmacies shipping to usa
https://mexicanpharmacy1st.shop/# mexican rx online
purple pharmacy mexico price list: buying prescription drugs in mexico – mexican drugstore online
mexico pharmacy mexico pharmacy buying prescription drugs in mexico
mexican mail order pharmacies: mexico drug stores pharmacies – mexican rx online
mexico pharmacies prescription drugs: mexico pharmacies prescription drugs – mexican pharmaceuticals online
https://mexicanpharmacy1st.online/# mexican border pharmacies shipping to usa
https://mexicanpharmacy1st.com/# buying prescription drugs in mexico
mexican mail order pharmacies mexico drug stores pharmacies mexican pharmacy
mexico pharmacy: mexico drug stores pharmacies – mexican rx online
buying prescription drugs in mexico: п»їbest mexican online pharmacies – mexican pharmaceuticals online
https://mexicanpharmacy1st.shop/# mexico pharmacies prescription drugs
mexico drug stores pharmacies medication from mexico pharmacy medicine in mexico pharmacies
mexican online pharmacies prescription drugs: mexican border pharmacies shipping to usa – reputable mexican pharmacies online
where to buy cheap clomid without rx: buy generic clomid without insurance – can you buy cheap clomid for sale
http://clomiphene.shop/# can i buy clomid prices
http://gabapentin.club/# generic neurontin 300 mg
can you order lisinopril online lisinopril metoprolol prinivil 20mg tabs
Abortion pills online: buy cytotec over the counter – buy cytotec in usa
can i buy generic clomid without rx: where can i buy cheap clomid without a prescription – can you get generic clomid pills
can i purchase generic clomid how to get generic clomid tablets can i purchase generic clomid pill
lisinopril 20 mg buy: lisinopril 10 mg online – lisinopril drug
order propecia propecia sale cost propecia no prescription
http://gabapentin.club/# price of neurontin
https://clomiphene.shop/# clomid without a prescription
32 neurontin: prescription medication neurontin – prescription drug neurontin
buying propecia without insurance cost cheap propecia price cost propecia no prescription
https://propeciaf.online/# cost generic propecia prices
order cytotec online: cytotec online – buy cytotec pills
https://propeciaf.online/# cost of propecia without prescription
cytotec buy online usa: Cytotec 200mcg price – п»їcytotec pills online
where to buy generic clomid: where can i buy clomid for sale – where to buy clomid online
https://propeciaf.online/# propecia cost
buy cytotec in usa Misoprostol 200 mg buy online cytotec abortion pill
http://clomiphene.shop/# can i get cheap clomid without insurance
buy propecia pill: buying generic propecia without dr prescription – cost cheap propecia without dr prescription
can i buy lisinopril over the counter zestril lisinopril lisinopril 5mg buy
how can i get clomid no prescription: how to get generic clomid for sale – can you buy generic clomid pill
cost propecia prices order propecia for sale order propecia pills
https://cytotec.xyz/# Cytotec 200mcg price
cost generic clomid without insurance: how can i get clomid tablets – where to get generic clomid pills
https://36and6health.shop/# online pharmacy prescription
buying prescription drugs in mexico pharmacies in mexico that ship to usa mexican mail order pharmacies
https://cheapestmexico.shop/# mexico drug stores pharmacies
https://36and6health.com/# online pharmacy prescription
mexican rx online: mexico drug stores pharmacies – mexico drug stores pharmacies
http://cheapestindia.com/# online pharmacy india
online shopping pharmacy india online pharmacy india indian pharmacy
https://cheapestindia.com/# Online medicine home delivery
https://cheapestandfast.shop/# no prescription pharmacy online
http://cheapestandfast.com/# canada prescription drugs online
pharmacy with no prescription: cheapest & fast pharmacy – medications online without prescriptions
http://cheapestindia.com/# п»їlegitimate online pharmacies india
https://cheapestcanada.shop/# canadian pharmacy in canada
online pharmacy canada no prescription cheapest & fast pharmacy mail order prescriptions from canada
http://cheapestmexico.com/# reputable mexican pharmacies online
https://cheapestcanada.com/# canadian medications
http://cheapestandfast.com/# no prescription needed pharmacy
pharmacie en ligne pas cher pharmacie en ligne france fiable Pharmacie sans ordonnance
farmacia online envГo gratis: farmacia online barata y fiable – farmacia online envГo gratis
Farmacia online più conveniente: Farmacia online miglior prezzo – farmacia online più conveniente
farmacia online: farmaci senza ricetta elenco – Farmacie on line spedizione gratuita
online apotheke rezept medikamente rezeptfrei internet apotheke
pharmacie en ligne livraison europe: pharmacie en ligne france – trouver un médicament en pharmacie
https://eufarmacieonline.com/# farmaci senza ricetta elenco
farmacie online sicure: farmaci senza ricetta elenco – acquistare farmaci senza ricetta
pharmacie en ligne france fiable pharmacie en ligne france pas cher п»їpharmacie en ligne france
farmacia online envÃo gratis: farmacia online envÃo gratis – farmacia online barata
medikament ohne rezept notfall: beste online-apotheke ohne rezept – eu apotheke ohne rezept
farmacia online: п»їFarmacia online migliore – acquistare farmaci senza ricetta
farmacia en casa online descuento: farmacia online barata y fiable – farmacias direct
farmacia online espaГ±a envГo internacional farmacia online madrid farmacia online 24 horas
pharmacie en ligne france livraison belgique: pharmacie en ligne france livraison belgique – pharmacie en ligne livraison europe
online apotheke preisvergleich: gГјnstige online apotheke – apotheke online
eu apotheke ohne rezept online apotheke internet apotheke
farmacia online españa: farmacias online seguras – farmacia online barata
https://eufarmaciaonline.shop/# farmacia online 24 horas
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
farmacia online barcelona: farmacia online madrid – farmacias online seguras
farmaci senza ricetta elenco comprare farmaci online con ricetta farmacie online autorizzate elenco
comprare farmaci online all’estero: comprare farmaci online all’estero – migliori farmacie online 2024
trouver un mГ©dicament en pharmacie: pharmacie en ligne fiable – pharmacie en ligne france livraison internationale
п»їFarmacia online migliore: farmacia online piГ№ conveniente – Farmacia online piГ№ conveniente
pharmacie en ligne sans ordonnance pharmacie en ligne pharmacie en ligne france livraison belgique
ohne rezept apotheke: europa apotheke – internet apotheke
pharmacie en ligne france pas cher: pharmacie en ligne – pharmacie en ligne france livraison internationale
https://eumedicamentenligne.com/# pharmacie en ligne sans ordonnance
online apotheke gГјnstig: online apotheke – medikament ohne rezept notfall
Farmacia online miglior prezzo migliori farmacie online 2024 farmacia online
farmacie online autorizzate elenco: acquistare farmaci senza ricetta – Farmacia online migliore
I would like to thank you for the efforts you have put in writing this blog. I’m hoping the same high-grade site post from you in the upcoming as well. Actually your creative writing abilities has inspired me to get my own website now. Really the blogging is spreading its wings rapidly. Your write up is a good example of it.
comprare farmaci online all’estero: Farmacia online miglior prezzo – farmacia online
comprare farmaci online con ricetta: farmacia online senza ricetta – migliori farmacie online 2024
farmacias online seguras: farmacia online barata – farmacia online envÃo gratis
pharmacie en ligne france livraison internationale pharmacie en ligne france livraison internationale Pharmacie en ligne livraison Europe
Hi there, just became alert to your blog through Google, and found that it’s truly informative. I?m gonna watch out for brussels. I will appreciate if you continue this in future. Lots of people will be benefited from your writing. Cheers!
pharmacie en ligne france livraison internationale: acheter mГ©dicament en ligne sans ordonnance – п»їpharmacie en ligne france
farmacia online senza ricetta: farmacia online più conveniente – comprare farmaci online all’estero
farmacia online madrid farmacia online 24 horas farmacias online seguras
https://euapothekeohnerezept.com/# ohne rezept apotheke
pharmacie en ligne avec ordonnance: cialis sans ordonnance – pharmacie en ligne pas cher
п»їpharmacie en ligne france: kamagra 100mg prix – pharmacie en ligne fiable
https://cenligne.com/# Pharmacie en ligne livraison Europe
acheter mГ©dicament en ligne sans ordonnance: kamagra oral jelly – pharmacie en ligne sans ordonnance
pharmacies en ligne certifiГ©es: cialis sans ordonnance – pharmacies en ligne certifiГ©es
trouver un mГ©dicament en pharmacie levitra generique sites surs pharmacie en ligne fiable
pharmacie en ligne france livraison internationale: pharmacie en ligne livraison europe – Pharmacie en ligne livraison Europe
Pharmacie en ligne livraison Europe: achat kamagra – pharmacie en ligne france livraison internationale
acheter mГ©dicament en ligne sans ordonnance: pharmacie en ligne pas cher – pharmacie en ligne avec ordonnance
Hello!
This post was created with XRumer 23 StrongAI.
Good luck 🙂
п»їpharmacie en ligne france: acheter mГ©dicament en ligne sans ordonnance – pharmacie en ligne fiable
Viagra sans ordonnance 24h suisse: Prix du Viagra en pharmacie en France – SildГ©nafil 100 mg prix en pharmacie en France
Viagra en france livraison rapide: Viagra generique en pharmacie – Viagra 100 mg sans ordonnance
п»їViagra sans ordonnance 24h: Viagra Pfizer sans ordonnance – Le gГ©nГ©rique de Viagra
This really answered my problem, thanks!
trouver un mГ©dicament en pharmacie: achat kamagra – п»їpharmacie en ligne france
Hello.
This post was created with XRumer 23 StrongAI.
Good luck 🙂
pharmacie en ligne pas cher: kamagra pas cher – pharmacie en ligne france fiable
https://phenligne.com/# pharmacie en ligne france fiable
One more thing. In my opinion that there are a lot of travel insurance websites of dependable companies that allow you to enter your vacation details and acquire you the estimates. You can also purchase the international holiday insurance policy on-line by using the credit card. All you have to do is to enter all travel information and you can be aware of the plans side-by-side. You only need to find the plan that suits your financial allowance and needs and use your bank credit card to buy the item. Travel insurance online is a good way to begin looking for a dependable company to get international travel cover. Thanks for giving your ideas.
pharmacie en ligne avec ordonnance: kamagra oral jelly – pharmacie en ligne france livraison internationale
pharmacie en ligne france pas cher: kamagra pas cher – Pharmacie Internationale en ligne
vente de mГ©dicament en ligne: acheter kamagra site fiable – pharmacie en ligne sans ordonnance
pharmacie en ligne sans ordonnance: levitra generique sites surs – pharmacie en ligne avec ordonnance
Pharmacie en ligne livraison Europe: levitra generique sites surs – pharmacie en ligne sans ordonnance
acheter mГ©dicament en ligne sans ordonnance: acheter kamagra site fiable – pharmacie en ligne france pas cher
Viagra vente libre pays: Acheter du Viagra sans ordonnance – Sildenafil teva 100 mg sans ordonnance
pharmacie en ligne pas cher: Levitra sans ordonnance 24h – п»їpharmacie en ligne france
acheter mГ©dicament en ligne sans ordonnance: Acheter Cialis 20 mg pas cher – Pharmacie sans ordonnance
Viagra homme prix en pharmacie: Meilleur Viagra sans ordonnance 24h – Quand une femme prend du Viagra homme
Pharmacie en ligne livraison Europe: pharmacie en ligne france livraison belgique – pharmacie en ligne pas cher
Pharmacie sans ordonnance: achat kamagra – pharmacie en ligne france fiable
Вы можете изменить свою жизнь.
ОПСУИМОЛОГ покажет вам, как это сделать.
Опсуимолог усвоил все, чему учит, на собственном горьком опыте: сначала испортив собственную жизнь и по необходимости открыв инструменты и исследования, которые изменили жизнь и привели его туда, где он есть сегодня.
Миссия Опсуимолога проста: поделиться проверенными инструментами, которые помогут вам создать лучшую жизнь. Загляните на сайт опсуимолога, и вы будете смеяться вместе, учиться и шаг за шагом создавать свою новую жизнь — свою лучшую жизнь.
Простые советы опсуимолога, подкрепленные исследованиями, изменили жизни множества людей, и теперь он раскрывает вам все свои трудные секреты, веселые неудачи и интересные истории, чтобы вы могли изменить свою.
Вы можете изменить свою жизнь.
ОПСУИМОЛОГ покажет вам, как это сделать.
Опсуимолог усвоил все, чему учит, на собственном горьком опыте: сначала испортив собственную жизнь и по необходимости открыв инструменты и исследования, которые изменили жизнь и привели его туда, где он есть сегодня.
Миссия Опсуимолога проста: поделиться проверенными инструментами, которые помогут вам создать лучшую жизнь. Загляните на сайт опсуимолога, и вы будете смеяться вместе, учиться и шаг за шагом создавать свою новую жизнь — свою лучшую жизнь.
Простые советы опсуимолога, подкрепленные исследованиями, изменили жизни множества людей, и теперь он раскрывает вам все свои трудные секреты, веселые неудачи и интересные истории, чтобы вы могли изменить свою.
Someone essentially assist to make critically posts I might state. That is the very first time I frequented your web page and so far? I surprised with the analysis you made to create this particular put up amazing. Fantastic activity!
https://autolux-azerbaijan.com/# Pin Up Azerbaycan
pin-up kazino: Pin Up Azerbaycan – Pin Up Azerbaycan ?Onlayn Kazino
https://autolux-azerbaijan.com/# Pin-up Giris
Pretty nice post. I just stumbled upon your weblog and wanted to say that I’ve really enjoyed browsing your weblog posts. After all I will be subscribing for your feed and I am hoping you write again very soon!
Pin-Up Casino: Pin up 306 casino – Pin-Up Casino
https://autolux-azerbaijan.com/# Pin Up
https://hasanhmt.com/ie-industrial-engineers
Pin-Up Casino: pin-up 141 casino – Pin Up Azerbaycan ?Onlayn Kazino
pin up 306: pin-up casino giris – pin up casino azerbaycan
Helpful information. Lucky me I found your website unintentionally, and I’m shocked why this twist of fate did not happened in advance! I bookmarked it.
pin up casino az https://azerbaijancuisine.com/# pin-up kazino
pin-up oyunu
I believe this web site has very great composed subject material posts.
medicine in mexico pharmacies: mexican pharmacy online – mexican drugstore online
https://northern-doctors.org/# buying from online mexican pharmacy
mexico pharmacy: mexican pharmacy – buying prescription drugs in mexico online
pharmacies in mexico that ship to usa mexican pharmacy online mexican pharmaceuticals online
http://northern-doctors.org/# buying from online mexican pharmacy
mexico drug stores pharmacies: mexican pharmacy online – purple pharmacy mexico price list
https://northern-doctors.org/# mexican rx online
F*ckin? amazing issues here. I am very satisfied to see your post. Thank you so much and i am having a look ahead to touch you. Will you please drop me a e-mail?
buying from online mexican pharmacy: mexican northern doctors – mexico pharmacies prescription drugs
buying prescription drugs in mexico: Mexico pharmacy that ship to usa – buying from online mexican pharmacy
medicine in mexico pharmacies mexican pharmacy mexican drugstore online
buying from online mexican pharmacy: Mexico pharmacy that ship to usa – buying prescription drugs in mexico online
http://northern-doctors.org/# buying prescription drugs in mexico
mexican rx online: northern doctors – mexico drug stores pharmacies
http://northern-doctors.org/# medicine in mexico pharmacies
I used to be suggested this blog by way of my cousin. I’m not positive whether or not this post is written by him as nobody else recognize such exact approximately my difficulty. You are incredible! Thanks!
reputable mexican pharmacies online: mexican pharmacy northern doctors – pharmacies in mexico that ship to usa
mexican pharmaceuticals online: mexican pharmacy northern doctors – mexican drugstore online
best online pharmacies in mexico northern doctors medication from mexico pharmacy
http://northern-doctors.org/# mexico pharmacies prescription drugs
reputable mexican pharmacies online: mexican pharmacy online – mexican mail order pharmacies
buying prescription drugs in mexico: northern doctors – mexican drugstore online
https://northern-doctors.org/# best online pharmacies in mexico
reputable mexican pharmacies online: Mexico pharmacy that ship to usa – mexico drug stores pharmacies
https://northern-doctors.org/# buying from online mexican pharmacy
reputable mexican pharmacies online: mexican northern doctors – medication from mexico pharmacy
medicine in mexico pharmacies Mexico pharmacy that ship to usa buying prescription drugs in mexico online
https://northern-doctors.org/# buying from online mexican pharmacy
medicine in mexico pharmacies: northern doctors pharmacy – mexico drug stores pharmacies
https://northern-doctors.org/# buying from online mexican pharmacy
mexico pharmacies prescription drugs: mexico drug stores pharmacies – mexico pharmacies prescription drugs
best online pharmacies in mexico Mexico pharmacy that ship to usa mexican mail order pharmacies
http://northern-doctors.org/# best online pharmacies in mexico
mexico pharmacies prescription drugs: northern doctors pharmacy – mexican drugstore online
mexican pharmaceuticals online: mexico drug stores pharmacies – mexico drug stores pharmacies
https://northern-doctors.org/# п»їbest mexican online pharmacies
mexico pharmacies prescription drugs: Mexico pharmacy that ship to usa – buying prescription drugs in mexico online
best online pharmacies in mexico: buying from online mexican pharmacy – mexican online pharmacies prescription drugs
https://northern-doctors.org/# reputable mexican pharmacies online
mexican pharmacy: mexican online pharmacies prescription drugs – mexican mail order pharmacies
http://northern-doctors.org/# purple pharmacy mexico price list
mexican border pharmacies shipping to usa: mexican northern doctors – п»їbest mexican online pharmacies
buying prescription drugs in mexico online Mexico pharmacy that ship to usa medication from mexico pharmacy
Увлекательный блокбастер Безумный Макс 2 – смотрите онлайн в превосходном качестве.
Насладитесь зрелищностью “Фуриоса: Хроники Безумного Макса” – просматривайте в сети в высоком качестве.
best online pharmacies in mexico: Mexico pharmacy that ship to usa – mexican rx online
reputable mexican pharmacies online: mexican pharmacy northern doctors – buying from online mexican pharmacy
https://northern-doctors.org/# purple pharmacy mexico price list
mexico pharmacy: northern doctors pharmacy – mexican drugstore online
https://northern-doctors.org/# buying prescription drugs in mexico online
mexican rx online: mexican northern doctors – buying prescription drugs in mexico
mexican pharmaceuticals online Mexico pharmacy that ship to usa pharmacies in mexico that ship to usa
reputable mexican pharmacies online: Mexico pharmacy that ship to usa – п»їbest mexican online pharmacies
https://northern-doctors.org/# purple pharmacy mexico price list
mexican drugstore online: mexican pharmacy northern doctors – buying prescription drugs in mexico
https://northern-doctors.org/# medication from mexico pharmacy
buying prescription drugs in mexico online: northern doctors – reputable mexican pharmacies online
buying prescription drugs in mexico online: mexican pharmacy northern doctors – mexican border pharmacies shipping to usa
pharmacies in mexico that ship to usa: mexican pharmacy – mexican drugstore online
buying from online mexican pharmacy northern doctors pharmacy mexican rx online
https://northern-doctors.org/# mexican drugstore online
purple pharmacy mexico price list cmq pharma best online pharmacies in mexico
http://cmqpharma.com/# medicine in mexico pharmacies
pharmacies in mexico that ship to usa
buying prescription drugs in mexico online cmq pharma buying prescription drugs in mexico
mexican pharmaceuticals online: mexico pharmacies prescription drugs – mexico pharmacy
purple pharmacy mexico price list cmqpharma.com pharmacies in mexico that ship to usa
mexican mail order pharmacies mexican pharmacy online п»їbest mexican online pharmacies
Can I simply say what a relief to seek out somebody who truly knows what theyre speaking about on the internet. You positively know how one can carry a difficulty to mild and make it important. More people must learn this and understand this aspect of the story. I cant believe youre not more popular because you definitely have the gift.
purple pharmacy mexico price list buying prescription drugs in mexico online mexican border pharmacies shipping to usa
medicine in mexico pharmacies
https://cmqpharma.com/# buying from online mexican pharmacy
buying from online mexican pharmacy
mexico drug stores pharmacies mexico drug stores pharmacies buying prescription drugs in mexico
mexico drug stores pharmacies medicine in mexico pharmacies buying from online mexican pharmacy
Shop authentic Marc Jacobs at unbeatable prices at the marc jacobs factory outlet online.
medication from mexico pharmacy buying prescription drugs in mexico pharmacies in mexico that ship to usa
One thing I’d really like to say is that car insurance termination is a horrible experience so if you’re doing the proper things like a driver you won’t get one. A number of people do obtain notice that they are officially dropped by their insurance company they have to scramble to get extra insurance after having a cancellation. Inexpensive auto insurance rates usually are hard to get after a cancellation. Knowing the main reasons regarding auto insurance termination can help drivers prevent burning off one of the most crucial privileges out there. Thanks for the suggestions shared through your blog.
Terrific work! That is the kind of info that are meant to be shared across the internet. Disgrace on the search engines for now not positioning this post higher! Come on over and discuss with my website . Thanks =)
I just couldn’t go away your website before suggesting that I extremely loved the usual information a person provide for your visitors? Is going to be back incessantly in order to investigate cross-check new posts
http://cmqpharma.com/# mexican border pharmacies shipping to usa
best online pharmacies in mexico
mexican rx online mexico pharmacy mexico drug stores pharmacies
Hi my loved one! I wish to say that this article is amazing, great written and come with almost all vital infos. I would like to see more posts like this .
Hi, I think your site might be having browser compatibility issues. When I look at your blog site in Opera, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, superb blog!
After examine a few of the blog posts in your website now, and I really like your means of blogging. I bookmarked it to my bookmark web site list and might be checking again soon. Pls take a look at my website as nicely and let me know what you think.
What?s Happening i’m new to this, I stumbled upon this I have discovered It positively useful and it has helped me out loads. I am hoping to give a contribution & assist different customers like its helped me. Good job.
I think this is among the most important info for me. And i am glad reading your article. But wanna remark on some general things, The site style is wonderful, the articles is really great : D. Good job, cheers
What?s Taking place i’m new to this, I stumbled upon this I’ve discovered It absolutely useful and it has helped me out loads. I hope to contribute & aid different customers like its helped me. Good job.
I’ve recently started a web site, the info you provide on this web site has helped me tremendously. Thanks for all of your time & work.
he blog was how do i say it… relevant, finally something that helped me. Thanks
It’s my belief that mesothelioma is definitely the most deadly cancer. It contains unusual properties. The more I really look at it the more I am certain it does not react like a true solid cells cancer. If perhaps mesothelioma is actually a rogue virus-like infection, therefore there is the potential for developing a vaccine plus offering vaccination to asbestos subjected people who are open to high risk with developing long term asbestos connected malignancies. Thanks for sharing your ideas for this important ailment.
Hello, i believe that i saw you visited my weblog thus i got here to “return the favor”.I am attempting to find things to improve my site!I guess its ok to make use of some of your concepts!!
mexico drug stores pharmacies
https://cmqpharma.com/# best online pharmacies in mexico
mexico drug stores pharmacies
Someone essentially help to make seriously posts I would state. This is the very first time I frequented your web page and thus far? I amazed with the research you made to create this particular publish amazing. Fantastic job!
Thanks for the suggestions you have discussed here. One more thing I would like to say is that personal computer memory requirements generally increase along with other developments in the technological innovation. For instance, when new generations of processor chips are introduced to the market, there’s usually a related increase in the shape demands of both computer system memory in addition to hard drive space. This is because the application operated by these processors will inevitably rise in power to make use of the new technological innovation.
Very nice post. I just stumbled upon your blog and wished to say that I’ve truly enjoyed browsing your blog posts. In any case I’ll be subscribing to your feed and I hope you write again very soon!
I discovered your blog site on google and check a few of your early posts. Continue to keep up the very good operate. I just additional up your RSS feed to my MSN News Reader. Seeking forward to reading more from you later on!…
Enjoyed reading this, very good stuff, thankyou. “Management is nothing more than motivating other people.” by Lee Iacocca.
Tonic Greens: An Overview Introducing Tonic Greens, an innovative immune support supplement meticulously crafted with potent antioxidants, essential minerals, and vital vitamins.
Thanks for the ideas you have shared here. Furthermore, I believe there are some factors which will keep your car insurance premium all the way down. One is, to contemplate buying automobiles that are inside the good list of car insurance organizations. Cars that are expensive are more at risk of being snatched. Aside from that insurance coverage is also in line with the value of your vehicle, so the higher priced it is, then higher your premium you only pay.
It’s a pity you don’t have a donate button! I’d certainly donate to this excellent blog! I suppose for now i’ll settle for book-marking and adding your RSS feed to my Google account. I look forward to fresh updates and will share this website with my Facebook group. Talk soon!
I like this web blog it’s a master piece!
Glad I discovered this ohttps://69v.topn google.Blog monry
This is a topic close to my heart cheers, where are your contact details though?
I’ll immediately grasp your rss feed as I can not in finding your email subscription hyperlink or newsletter service. Do you have any? Kindly permit me understand so that I may subscribe. Thanks.
I have been absent for a while, but now I remember why I used to love this blog. Thank you, I?¦ll try and check back more often. How frequently you update your site?
I besides think therefore, perfectly indited post! .
Thank you, I have recently been searching for info about this subject matter for ages and yours is the best I’ve found so far.
I got what you mean , thankyou for posting.Woh I am lucky to find this website through google. “Being intelligent is not a felony, but most societies evaluate it as at least a misdemeanor.” by Lazarus Long.
I dugg some of you post as I thought they were invaluable very helpful
Regards for helping out, excellent information.
Definitely, what a splendid site and revealing posts, I definitely will bookmark your website.All the Best!
Porn
Buy Drugs
As a Newbie, I am constantly browsing online for articles that can help me. Thank you
Some genuinely interesting details you have written.Aided me a lot, just what I was searching for : D.
Viagra
Buy Drugs
Viagra
Viagra
Scam
Pornstar
Buy Drugs
Scam
Viagra
Porn
Pornstar
Hey very nice website!! Man .. Excellent .. Wonderful .. I will bookmark your website and take the feeds additionallyKI’m happy to seek out so many useful information right here within the submit, we want work out extra techniques in this regard, thank you for sharing. . . . . .
Pornstar
Porn site
I envy your work, thanks for all the useful content.
Pornstar
Sex
Pornstar
As a Newbie, I am always searching online for articles that can help me. Thank you
F*ckin’ amazing things here. I am very glad to see your post. Thanks a lot and i’m looking forward to contact you. Will you please drop me a e-mail?
whoah this blog is wonderful i love reading your articles. Keep up the good work! You know, a lot of people are hunting around for this info, you can aid them greatly.
Porn site
Buy Drugs
Pornstar
Porn site
Viagra
Porn site
Pornstar
Viagra
Thanks for any other fantastic article. The place else may just anyone get that kind of info in such an ideal approach of writing? I have a presentation next week, and I am on the search for such information.
Pornstar
Scam
I truly enjoy looking through on this web site, it contains great blog posts.
My brother recommended I might like this blog. He was totally right. This post actually made my day. You can not imagine simply how much time I had spent for this information! Thanks!
I do love the manner in which you have presented this particular situation plus it does indeed give us a lot of fodder for thought. On the other hand, coming from everything that I have experienced, I only trust as other responses stack on that men and women continue to be on issue and not get started on a soap box associated with some other news of the day. Still, thank you for this outstanding point and while I can not agree with it in totality, I respect the standpoint.
This is very interesting, You are a very skilled blogger. I’ve joined your rss feed and look forward to seeking more of your fantastic post. Also, I have shared your web site in my social networks!
Viagra
Porn
Great line up. We will be linking to this great article on our site. Keep up the good writing.
Thanks for helping out, great information.
Buy Drugs
Pornstar
Buy Drugs
Scam
Perfectly written content, thanks for entropy. “Life is God’s novel. Let him write it.” by Isaac Bashevis Singer.
Viagra
Viagra
Scam
Viagra
Porn
Buy Drugs
Viagra
Sex
Porn site
Pornstar
https://foruspharma.com/# mexico drug stores pharmacies
canadian pharmacy com buying drugs from canada canada drugs reviews
https://foruspharma.com/# п»їbest mexican online pharmacies
buying prescription drugs in mexico online: buying prescription drugs in mexico online – mexican rx online
india pharmacy mail order buy prescription drugs from india india pharmacy mail order
purple pharmacy mexico price list: mexican mail order pharmacies – mexican pharmaceuticals online
indianpharmacy com: cheapest online pharmacy india – top 10 pharmacies in india
Viagra
Scam
pharmacies in mexico that ship to usa: pharmacies in mexico that ship to usa – mexico drug stores pharmacies
Pornstar
Sex
Porn
reputable indian online pharmacy: reputable indian online pharmacy – indian pharmacy
Scam
global pharmacy canada canadian pharmacy store safe reliable canadian pharmacy
medicine in mexico pharmacies: pharmacies in mexico that ship to usa – reputable mexican pharmacies online
Porn site
Porn site
safe online pharmacies in canada best canadian pharmacy to order from canadian drugs
https://indiapharmast.com/# best online pharmacy india
india online pharmacy: Online medicine home delivery – indian pharmacy online
п»їlegitimate online pharmacies india: best online pharmacy india – mail order pharmacy india
mexican pharmacy: reputable mexican pharmacies online – medicine in mexico pharmacies
https://indiapharmast.com/# best online pharmacy india
I discovered your blog site on google and check a few of your early posts. Continue to keep up the very good operate. I just additional up your RSS feed to my MSN News Reader. Seeking forward to reading more from you later on!…
Found your site within the yahoo bulk shirts warehouse directory, very nice job, thanks Screen printing could further be used for those types of other substrates from plastic to metal Despite the fact that tiny and complex details can be gathered, screen printing is preferably suitable for bold and graphic designs
mexican drugstore online: mexican drugstore online – mexican drugstore online
Buy Drugs
Sex
As soon as I discovered this internet site I went on reddit to share some of the love with them.
reputable indian pharmacies best online pharmacy india pharmacy website india
indianpharmacy com: indian pharmacy online – cheapest online pharmacy india
mexican pharmaceuticals online mexican pharmacy п»їbest mexican online pharmacies
canadian world pharmacy: canada drugstore pharmacy rx – canadian pharmacy 1 internet online drugstore
https://indiapharmast.com/# indian pharmacy paypal
purple pharmacy mexico price list: mexican online pharmacies prescription drugs – pharmacies in mexico that ship to usa
canadian medications: rate canadian pharmacies – pharmacy canadian superstore
mexican border pharmacies shipping to usa: medicine in mexico pharmacies – buying prescription drugs in mexico
https://canadapharmast.com/# online canadian pharmacy review
canadian pharmacy victoza canadian pharmacy 365 onlinecanadianpharmacy 24
Online medicine order: india pharmacy mail order – online pharmacy india
Sex
Scam
safe canadian pharmacy: canadian pharmacies comparison – canadian pharmacy 365
You have noted very interesting points! ps decent internet site.
Buy Drugs
Porn
Viagra
Sex
top online pharmacy india: Online medicine order – reputable indian online pharmacy
Buy Drugs
What i do not realize is actually how you’re not really a lot more neatly-liked than you may be now. You’re very intelligent. You realize therefore significantly in relation to this topic, made me personally believe it from numerous numerous angles. Its like women and men don’t seem to be involved except it’s something to do with Woman gaga! Your individual stuffs outstanding. All the time handle it up!
reputable mexican pharmacies online: mexican rx online – best online pharmacies in mexico
mexican drugstore online: mexico pharmacies prescription drugs – purple pharmacy mexico price list
It?s actually a cool and helpful piece of information. I?m happy that you simply shared this useful information with us. Please stay us informed like this. Thanks for sharing.
magnificent post, very informative. I wonder why the other specialists of this sector don’t notice this. You must continue your writing. I’m confident, you’ve a huge readers’ base already!
Sex
Very interesting subject, thankyou for posting. “Experience a comb life gives you after you lose your hair.” by Judith Stern.
buy cipro without rx: buy cipro online – buy cipro online without prescription
Sex
Pornstar
buy cipro without rx: buy generic ciprofloxacin – ciprofloxacin
http://clomiddelivery.pro/# where to get cheap clomid without a prescription
http://paxloviddelivery.pro/# buy paxlovid online
Sex
I have been absent for a while, but now I remember why I used to love this website. Thank you, I will try and check back more often. How frequently you update your website?
http://amoxildelivery.pro/# over the counter amoxicillin canada
Very great post. I simply stumbled upon your blog and wished to say that I have really loved browsing your blog posts. After all I will be subscribing to your feed and I hope you write again soon!
paxlovid pharmacy: Paxlovid over the counter – paxlovid cost without insurance
http://amoxildelivery.pro/# cost of amoxicillin 30 capsules
Write more, thats all I have to say. Literally, it seems as though you relied on the video to make your point. You clearly know what youre talking about, why throw away your intelligence on just posting videos to your weblog when you could be giving us something enlightening to read?
buy ciprofloxacin over the counter: buy cipro online canada – buy cipro without rx
Sex
Sex
Porn
Scam
Scam
Viagra
Porn
Viagra
http://clomiddelivery.pro/# where can i get generic clomid without insurance
http://amoxildelivery.pro/# canadian pharmacy amoxicillin
cipro ciprofloxacin: buy cipro online – ciprofloxacin generic price
I visited a lot of website but I think this one holds something special in it in it
cost of clomid without dr prescription: where can i get cheap clomid pills – clomid rx
Porn site
Pornstar
https://paxloviddelivery.pro/# paxlovid for sale
Sex
It is the best time to make some plans for the longer term and it is time to be happy. I have read this put up and if I may I desire to counsel you some attention-grabbing things or advice. Perhaps you can write subsequent articles referring to this article. I wish to read more things approximately it!
http://amoxildelivery.pro/# amoxicillin 500mg pill
I like gathering utile information , this post has got me even more info! .
https://ciprodelivery.pro/# purchase cipro
Sex
Viagra
Sex
Sex
https://paxloviddelivery.pro/# buy paxlovid online
Wonderful work! This is the type of info that should be shared around the internet. Shame on the search engines for not positioning this post higher! Come on over and visit my site . Thanks =)
order generic clomid without rx: clomid rx – how can i get generic clomid without insurance
Buy Drugs
Scam
can i get clomid without rx: get cheap clomid price – cost generic clomid without dr prescription
Also a thing to mention is that an online business administration training is designed for scholars to be able to efficiently proceed to bachelors degree education. The Ninety credit certification meets the other bachelor education requirements and when you earn your associate of arts in BA online, you may have access to the newest technologies in this particular field. Several reasons why students would like to get their associate degree in business is because they’re interested in the field and want to find the general instruction necessary ahead of jumping into a bachelor college diploma program. Thanks alot : ) for the tips you actually provide inside your blog.
Viagra
Viagra
Pornstar
Porn site
https://paxloviddelivery.pro/# п»їpaxlovid
http://doxycyclinedelivery.pro/# buy doxycycline online usa
This actually answered my problem, thank you!
http://amoxildelivery.pro/# buy amoxicillin online without prescription
Paxlovid buy online: Paxlovid buy online – paxlovid for sale
https://clomiddelivery.pro/# can i order generic clomid without dr prescription
ciprofloxacin order online: buy cipro without rx – ciprofloxacin 500 mg tablet price
Your article helped me a lot, is there any more related content? Thanks!
https://clomiddelivery.pro/# where can i get clomid now
You really make it seem so easy with your presentation but I find this topic to be actually something which I think I would never understand. It seems too complicated and very broad for me. I’m looking forward for your next post, I will try to get the hang of it!
https://ciprodelivery.pro/# cipro ciprofloxacin
Enjoyed reading this, very good stuff, regards.
Buy Drugs
Sex
Scam
can i buy clomid without rx: how to get generic clomid no prescription – can i get cheap clomid pills
Porn
Porn site
Porn site
buy cipro: buy cipro without rx – cipro
http://ciprodelivery.pro/# antibiotics cipro
Buy Drugs
Viagra
Porn
Hello. remarkable job. I did not imagine this. This is a splendid story. Thanks!
Viagra
Porn site
Porn
Buy Drugs
canadian pharmacy amoxicillin: amoxicillin in india – amoxicillin 250 mg
doxycycline 100mg cost uk: 2985 doxycycline – doxycycline 75 mg capsules
This is very interesting, You are a very skilled blogger. I’ve joined your feed and look forward to seeking more of your wonderful post. Also, I’ve shared your website in my social networks!
Viagra
Scam
I?¦ve been exploring for a little for any high quality articles or blog posts in this sort of space . Exploring in Yahoo I at last stumbled upon this site. Reading this info So i?¦m happy to show that I’ve a very excellent uncanny feeling I came upon just what I needed. I so much definitely will make sure to do not disregard this site and give it a glance on a constant basis.
Buy Drugs
Porn
Viagra
Really instructive and excellent structure of articles, now that’s user friendly (:.
Porn
Buy Drugs
Hiya, I’m really glad I’ve found this information. Nowadays bloggers publish just about gossips and net and this is really irritating. A good blog with interesting content, this is what I need. Thanks for keeping this web-site, I will be visiting it. Do you do newsletters? Cant find it.
I have been browsing online more than three hours these days, yet I never found any fascinating article like yours. It is beautiful price enough for me. Personally, if all web owners and bloggers made good content material as you probably did, the net will probably be a lot more useful than ever before.
Great tremendous things here. I?¦m very glad to peer your post. Thanks so much and i’m looking forward to contact you. Will you kindly drop me a mail?
Sex
Porn site
Scam
Pornstar
It’s best to participate in a contest for one of the best blogs on the web. I’ll suggest this site!
Sex
Scam
There is noticeably a bundle to know about this. I assume you made certain nice points in features also.
Porn
Sex
Great post. I was checking continuously this blog and I’m impressed! Very useful information particularly the last part 🙂 I care for such info a lot. I was looking for this certain info for a very long time. Thank you and good luck.
Pretty! This was a really wonderful post. Thank you for your provided information.
Great post, you have pointed out some fantastic points, I also believe this s a very good website.